A closer look at the most common gene mutations in ALS*
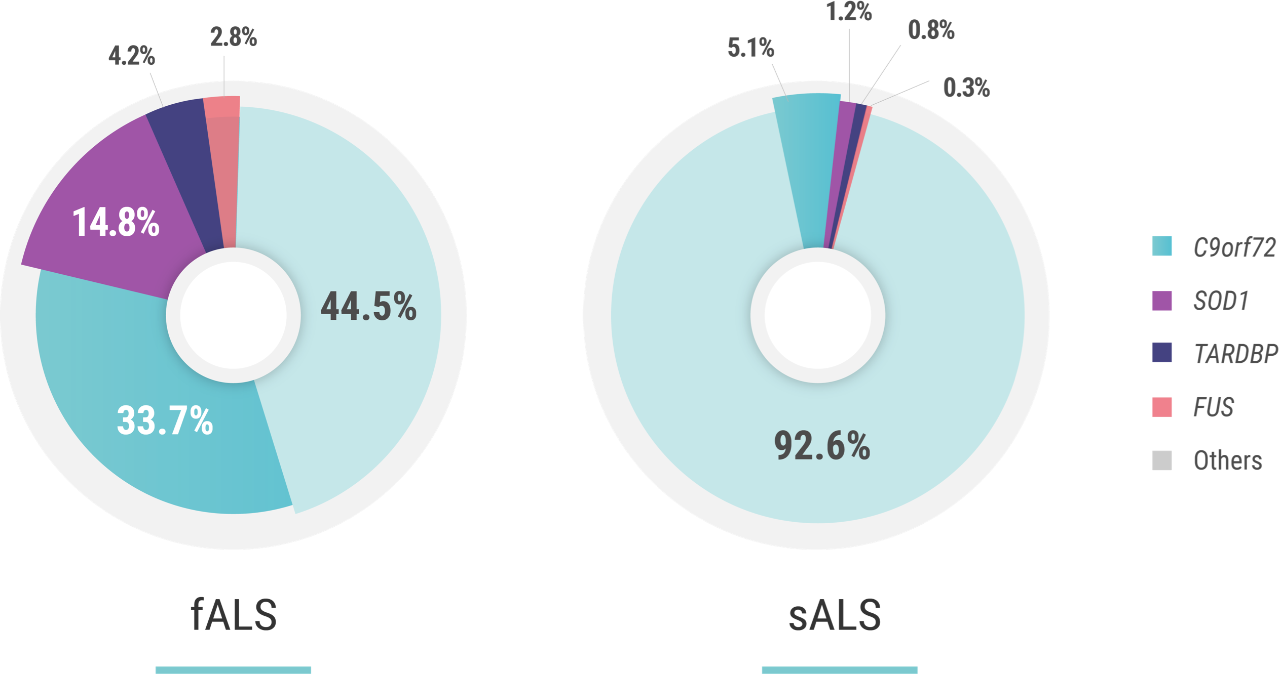
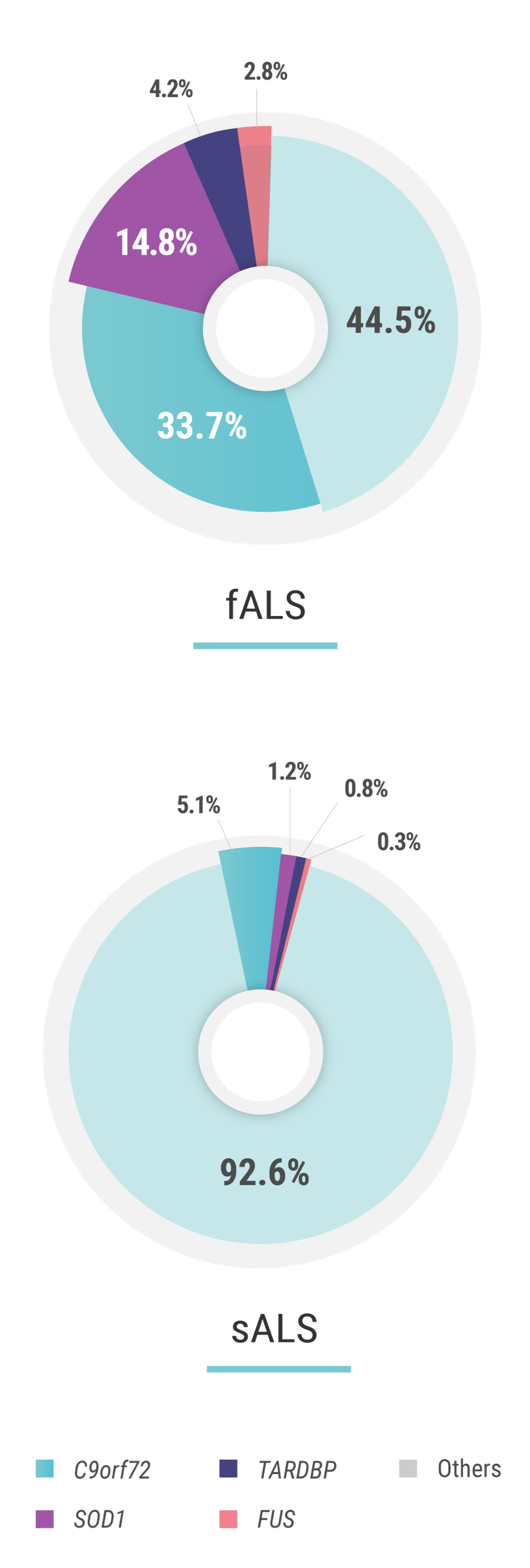
Several genes have been identified as having a strong association with ALS.2,3 The first, discovered in 1993, is SOD1 (superoxide dismutase 1),4 which accounts for ~15% of fALS (familial ALS) cases and can be found in ~1% of sALS (sporadic ALS) cases.2 C9orf72 (chromosome 9 open reading frame 72) expansion mutations, discovered in 2011,5 are the most common, and account for nearly 33% of identified fALS cases and have been found in ~5% of sALS patients.2,5
The next most common mutations associated with ALS are found in the TARDBP (TAR DNA binding protein) and FUS (fused in sarcoma) genes. Mutations in these genes each account for approximately 3%-4% of fALS cases and approximately 1% of sALS cases.1 The discovery and investigation of these genes hold great promise for a better understanding of genetic ALS.
*Based on global population data.
C9orf72-ALS
Although more patients with genetic ALS carry C9orf72 mutations than any other type globally, frequencies vary greatly by ethnicity and geographic region. For example, the highest frequencies of C9orf72 expansion mutations are found in Scandinavian countries, while SOD1 mutations are much more common in Asia.1,6,7
SOD1-ALS
In patients with SOD1-ALS, the SOD1 gene has undergone a change, or mutation, which results in damage to nerves in the brain and spinal cord that control muscle movement. SOD1 gene mutations account for just 2% of all ALS cases—but 20% of those with a genetic component.1 Nearly half of all patients living with SOD1-ALS, do not have a known family history of the disease.8-11
TARDBP-ALS
Mutations in the TARDBP gene are found in approximately 4% of patients with fALS and 1% of patients with sALS.1,9 While these mutations are believed to be less common than those in the SOD1 and C9orf72 genes, TARDBP mutations have been identified in patients from a diverse group of countries and geographical regions, suggesting that these mutations may be a factor in who develops genetic ALS worldwide, being identified in Japan, Australia, North America, and China.1,9
FUS-ALS
ALS-associated FUS mutations were first reported in 2009 and are now found in approximately 4% of patients with fALS and less than 1% of patients with sALS.1,9 While the course of disease among patients with FUS-ALS can be unpredictable, certain FUS mutations have also been linked to an earlier onset of genetic ALS.1,9 FUS mutations have been identified among patients with genetic ALS in a wide range of geographic regions, including other parts of Europe, Australia, North America, Asia, and Africa.9
References: 1. Roggenbuck J, Quick A, Kolb SJ. Genetic testing and genetic counseling for amyotrophic lateral sclerosis: an update for clinicians. Genet Med. 2017;19(3):267-274. 2. Zou ZY, Zhou ZR, Che CH, et al. Genetic epidemiology of amyotrophic lateral sclerosis: a systematic review and meta-analysis. J Neurol Neurosurg Psychiatry. 2017;88(7):540-549. 3. Bali T, Self W, Liu J, et al. Defining SOD1 ALS natural history to guide therapeutic clinical trial design. J Neurol Neurosurg Psychiatry. 2017;88(2):99-105. 4. Rosen DR. Mutations in Cu/Zn superoxide dismutase gene are associated with familial amyotrophic lateral sclerosis. Nature.1993;364(6435):362. 5. Balendra R, Isaacs AM. C9orf72-mediated ALS and FTD: multiple pathways to disease. Nat Rev Neurol. 2018;14(9):544-588. doi:10.1038/s41582-018-0047-2 6. Wei Q, Zhou Q, Chen Y, et al. Analysis of SOD1 mutations in a Chinese population with amyotrophic lateral sclerosis: a case-control study and literature review. Sci Rep. 2017;7:44606. doi:10.1038/srep44606 7. Shahrizaila N, Sobue G, Kuwabara S, et al. Amyotrophic lateral sclerosis and motor neuron syndromes in Asia. J Neurol Neurosurg Psychiatry. 2016;87:821-830. 8. Chiò A, Mazzini L, D’Alfonso S, et al. The multistep hypothesis of ALS revisited: the role of genetic mutations. Neurology. 2018;91(7):e635-e642. doi:10.1212/WNL.0000000000005996 9. Lattante S, Marangi G, Doronzio PN, et al. High-throughput genetic testing in ALS: the challenging path of variant classification considering the ACMG guidelines. Genes (Basel). 2020;11(10):1123. doi:10.3390/genes11101123 10. Arthur KC, Calvo A, Price TR, et al. Projected increase in amyotrophic lateral sclerosis from 2015 to 2040. Nat Commun. 2016;7:12408. 11. Zarei S, Carr K, Reiley L, et al. A comprehensive review of amyotrophic lateral sclerosis. Surg Neurol Int. 2015;6:171.
1. Roggenbuck J, Quick A, Kolb SJ. Genetic testing and genetic counseling for amyotrophic lateral sclerosis: an update for clinicians. Genet Med. 2017;19(3):267-274. 2. Zou ZY, Zhou ZR, Che CH, et al. Genetic epidemiology of amyotrophic lateral sclerosis: a systematic review and meta-analysis. J Neurol Neurosurg Psychiatry. 2017;88(7):540-549. 3. Bali T, Self W, Liu J, et al. Defining SOD1 ALS natural history to guide therapeutic clinical trial design. J Neurol Neurosurg Psychiatry. 2017;88(2):99-105. 4. Rosen DR. Mutations in Cu/Zn superoxide dismutase gene are associated with familial amyotrophic lateral sclerosis. Nature.1993;364(6435):362. 5. Balendra R, Isaacs AM. C9orf72-mediated ALS and FTD: multiple pathways to disease. Nat Rev Neurol. 2018;14(9):544-588. doi:10.1038/s41582-018-0047-2 6. Wei Q, Zhou Q, Chen Y, et al. Analysis of SOD1 mutations in a Chinese population with amyotrophic lateral sclerosis: a case-control study and literature review. Sci Rep. 2017;7:44606. doi:10.1038/srep44606 7. Shahrizaila N, Sobue G, Kuwabara S, et al. Amyotrophic lateral sclerosis and motor neuron syndromes in Asia. J Neurol Neurosurg Psychiatry. 2016;87:821-830. 8. Chiò A, Mazzini L, D’Alfonso S, et al. The multistep hypothesis of ALS revisited: the role of genetic mutations. Neurology. 2018;91(7):e635-e642. doi:10.1212/WNL.0000000000005996 9. Lattante S, Marangi G, Doronzio PN, et al. High-throughput genetic testing in ALS: the challenging path of variant classification considering the ACMG guidelines. Genes (Basel). 2020;11(10):1123. doi:10.3390/genes11101123 10. Arthur KC, Calvo A, Price TR, et al. Projected increase in amyotrophic lateral sclerosis from 2015 to 2040. Nat Commun. 2016;7:12408. 11. Zarei S, Carr K, Reiley L, et al. A comprehensive review of amyotrophic lateral sclerosis. Surg Neurol Int. 2015;6:171.