Genetic ALS:
an overview of
common mutations
Genetic ALS / An overview of common mutations
SOD1: the “pioneer” genetic mutation
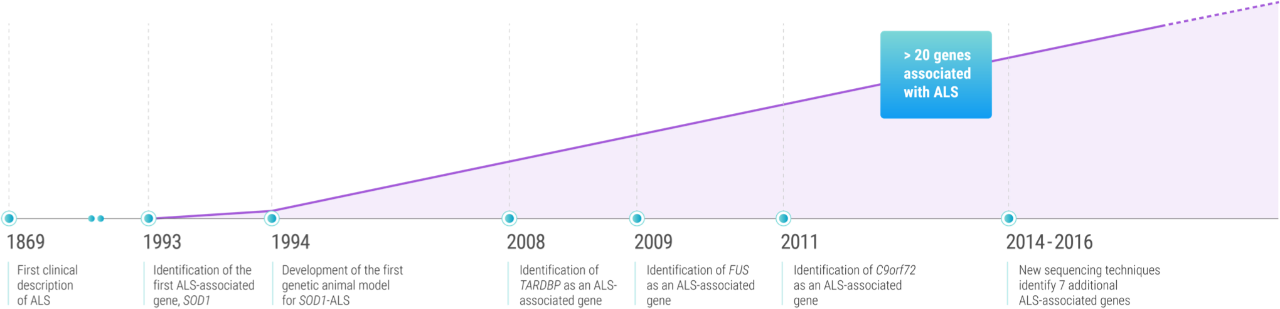
What we now call genetic ALS was first discovered in 1993, when Robert Brown’s research group identified 11 different SOD1 mutations in familial cases of ALS.1,2
Researchers would eventually come to understand that mutant SOD1 proteins can cause excitotoxicity in motor neurons, due to failure in clearing neurotransmitters that trigger motor activity. Over time, hyperexcitable neurons gradually become more vulnerable to motor neuron death, causing the progressive muscle atrophy, spasticity, and fatal respiratory decline seen in ALS.4,5
SOD1 mutations are the second most common mutations among patients with genetic ALS.5 Approximately 187 SOD1 variants have been described in both familial and sporadic cases of ALS to date.
Currently, up to 20% of familial cases (fALS) and 3% of sporadic cases (sALS) have been linked to SOD1 mutations, and these numbers are increasing as systematic genetic screening methods are incorporated into clinical practice.1,6,7


Considerable phenotypic heterogeneity is seen across different SOD1 variants.
For example, the A4V mutation, which is the most frequent variant in North America, gives rise to a fast-progressing form of ALS that typically leads to death within one year from symptom onset. In contrast, the homozygous D90A mutation is associated with an indolent disease course, with patients reaching respiratory failure after 10 or more years of illness.8
SOD1-ALS also manifests in a heterogeneous manner in different parts of the world. The highest frequencies are reported in patients in Japan, Taiwan, and Korea.6
While mutations in the SOD1 gene were the first to be linked to ALS,5 they were hardly the last to help researchers in their pursuit of a deeper understanding of this devastating disease.
Key milestones in genetic ALS2,5,9-12
C9orf72: the most common ALS-associated mutation
First identified in 2011, C9orf72 repeat expansion mutations may not have been among the earliest to be associated with ALS, but they are by far the most common. A pathogenic hexanucleotide (G4C2) repeat expansion in the first intron of C9orf72 accounts for approximately 40% of fALS and 7% of sALS cases. The C9orf72 repeat expansion is the only mutation found in more than 2% of patients with sALS.5
Three mechanisms have been proposed for how G4C2 hexanucleotide repeat expansion (HRE) in C9orf72 may induce neurodegenerative changes, including loss of C9orf72 function through haploinsufficiency, toxic gain-of-function due to the generation of aberrant HRE-containing RNA, and toxic gain-of-function through the accumulation of dipeptide repeat proteins translated from hexanucleotide repeat RNA.13
C9orf72 repeat expansion mutations are transmitted in an autosomal-dominant fashion, with variable and age-dependent penetrance. Studies have estimated cumulative age-dependent penetrance of 0% at age 35, 50% at 58, and almost 100% by 80.5
In addition to the upper and lower motor neuron dysfunction common with ALS, C9orf72 repeat expansion mutations have been linked to frontotemporal lobar degeneration that manifests clinically with behavior changes, executive dysfunction, and language impairment.5
The frequency of C9orf72 expansion mutations varies greatly by ethnicity and geography, with the highest frequencies reported in northern Europe and the lowest frequencies reported in Asia.5
TARDBP and FUS: mutations found around the world
Following C9orf72 and SOD1 mutations, the most common mutations associated with ALS are in the TARDBP and FUS genes. Mutations in these genes are found in approximately 4% of patients with fALS and 1% of patients with sALS. However, researchers have noted that more studies are needed to determine the true frequency of TARDBP mutations among individuals with sALS.5,12
First linked to ALS in 2008,11 TARDBP mutations have been identified across a diverse group of geographical regions, suggesting that they may be a factor in who develops genetic ALS worldwide. While TARDBP-ALS is most prevalent in Europe, ALS-associated TARDBP mutations have been identified in Japan, Australia, North America, and China.12
ALS-associated FUS mutations were first reported in 2009.12 While the course of disease among FUS-ALS patients is highly variable, patients with these mutations are known to have shorter lifespans than those with other mutations.5 A number of FUS mutations have been linked to an earlier onset of genetic ALS, with some patients showing symptoms before the age of 25.12
As with TARDBP mutations, FUS mutations have been identified among patients with genetic ALS across geographic regions, including Europe, Australia, North America, Asia, and Africa.12
New technologies, new discoveries
While researchers can’t be certain how many ALS-associated mutations remain to be identified, new technologies have accelerated the pace of discovery in recent years.11
Since 2014, new sequencing technologies—including genome-wide association studies, whole genome studies, and exome sequencing—have led to the discovery of ALS-associated mutations in 7 additional genes: MATR3, CHCHD10, TBK1, TUBA4A, NEK1, C21orf2, and CCNF.11
As with many common ALS-associated mutations, these mutations have been found in patients with fALS as well as sALS11—making clear once again that genetic ALS can affect those with or without a family history of the disease.
References: 1. Kim G, Gautier O, Tassoni-Tsuchida E, Ma XR, Gitler AD. ALS genetics: gains, losses, and implications for future therapies. Neuron. 2020;108(5):822-842. doi:10.1016/j.neuron.2020.08.02. 2. Rosen DR. Mutations in Cu/Zn superoxide dismutase gene are associated with familial amyotrophic lateral sclerosis. Nature. 1993;364(6435):362. doi:10.1038/364362c0 3. Laferriere F, Polymenidou M. Advances and challenges in understanding the multifaceted pathogenesis of amyotrophic lateral sclerosis. Swiss Med Wkly. 2015;145:w14054. doi:10.4414/smw.2015.14054 4. McCann EP, Williams KL, Fifita JA, et al. The genotype-phenotype landscape of familial amyotrophic lateral sclerosis in Australia. Clin Genet. 2017;92(3):259-266. 5. Roggenbuck J, Quick A, Kolb SJ. Genetic testing and genetic counseling for amyotrophic lateral sclerosis: an update for clinicians. Genet Med. 2017;19(3):267-274. 6. Nakamura R, Sone J, Atsuta N, et al. Next-generation sequencing of 28 ALS-related genes in a Japanese ALS cohort. Neurobiol Aging. 2016;39:219.e1-219.e2198. doi:10.1016/j.neurobiolaging.2015.11.030 7. Shepheard SR, Parker MD, Cooper-Knock J, et al; on behalf of Project MINE Consortium; Project MinE. Value of systematic genetic screening of patients with amyotrophic lateral sclerosis. J Neurol Neurosurg Psychiatry. 2021;92(5):510-518. 8. Renton AE, Chiò A, Traynor BJ. State of play in amyotrophic lateral sclerosis genetics. Nat Neurosci. 2014;17(1):17-23. 9. Masrori P, Van Damme P. Amyotrophic lateral sclerosis: a clinical review. Eur J Neurol. 2020;27(10):1918-1929. 10. Picher-Martel V, Valdmanis PN, Gould PV, Julien JP, Dupré N. From animal models to human disease: a genetic approach for personalized medicine in ALS. Acta Neuropathol Commun. 2016;4(1):70. doi:10.1186/s40478-016-0340-5 11. Chia R, Chiò A, Traynor BJ. Novel genes associated with amyotrophic lateral sclerosis: diagnostic and clinical implications. Lancet Neurol. 2018;17(1):94-102. doi:10.1016/S1474-4422(17)30401-5 12. Lattante S, Rouleau GA, Kabashi E. TARDBP and FUS mutations associated with amyotrophic lateral sclerosis: summary and update. Hum Mutat. 2013;34(6):812-826. 13. Leko MB, Župunski V, Kirincich J, et al. Molecular mechanisms of neurodegeneration related to C9orf72 hexanucleotide repeat expansion. Behav Neurol. 2019:2909168. doi:10.1155/2019/2909168
1. Kim G, Gautier O, Tassoni-Tsuchida E, Ma XR, Gitler AD. ALS genetics: gains, losses, and implications for future therapies. Neuron. 2020;108(5):822-842. doi:10.1016/j.neuron.2020.08.02. 2. Rosen DR. Mutations in Cu/Zn superoxide dismutase gene are associated with familial amyotrophic lateral sclerosis. Nature. 1993;364(6435):362. doi:10.1038/364362c0 3. Laferriere F, Polymenidou M. Advances and challenges in understanding the multifaceted pathogenesis of amyotrophic lateral sclerosis. Swiss Med Wkly. 2015;145:w14054. doi:10.4414/smw.2015.14054 4. McCann EP, Williams KL, Fifita JA, et al. The genotype-phenotype landscape of familial amyotrophic lateral sclerosis in Australia. Clin Genet. 2017;92(3):259-266. 5. Roggenbuck J, Quick A, Kolb SJ. Genetic testing and genetic counseling for amyotrophic lateral sclerosis: an update for clinicians. Genet Med. 2017;19(3):267-274. 6. Nakamura R, Sone J, Atsuta N, et al. Next-generation sequencing of 28 ALS-related genes in a Japanese ALS cohort. Neurobiol Aging. 2016;39:219.e1-219.e2198. doi:10.1016/j.neurobiolaging.2015.11.030 7. Shepheard SR, Parker MD, Cooper-Knock J, et al; on behalf of Project MINE Consortium; Project MinE. Value of systematic genetic screening of patients with amyotrophic lateral sclerosis. J Neurol Neurosurg Psychiatry. 2021;92(5):510-518. 8. Renton AE, Chiò A, Traynor BJ. State of play in amyotrophic lateral sclerosis genetics. Nat Neurosci. 2014;17(1):17-23. 9. Masrori P, Van Damme P. Amyotrophic lateral sclerosis: a clinical review. Eur J Neurol. 2020;27(10):1918-1929. 10. Picher-Martel V, Valdmanis PN, Gould PV, Julien JP, Dupré N. From animal models to human disease: a genetic approach for personalized medicine in ALS. Acta Neuropathol Commun. 2016;4(1):70. doi:10.1186/s40478-016-0340-5 11. Chia R, Chiò A, Traynor BJ. Novel genes associated with amyotrophic lateral sclerosis: diagnostic and clinical implications. Lancet Neurol. 2018;17(1):94-102. doi:10.1016/S1474-4422(17)30401-5 12. Lattante S, Rouleau GA, Kabashi E. TARDBP and FUS mutations associated with amyotrophic lateral sclerosis: summary and update. Hum Mutat. 2013;34(6):812-826. 13. Leko MB, Župunski V, Kirincich J, et al. Molecular mechanisms of neurodegeneration related to C9orf72 hexanucleotide repeat expansion. Behav Neurol. 2019:2909168. doi:10.1155/2019/2909168